|
|
|
GPCR Peptide Ligand Library (L-004)
|
|
a collection of nature-inspired design of domain-specific peptides |
This library is being continuously updated to include all new and current peptides including GnRH(1-5) (GPR101 and GPR173), SDF-1-alpha (CXCR4 receptor), Ela-32 and Ela-21 (APJ), SCNH2/Apelin-prepropeptide(42-57), TLQP-21 (C3a or C1q receptor), D-[Lys3]GHRP-6 (CCR5), Neuronostatin (GPR107), Pepcan-12 (CB1) Fractalkine peptides, cysteine-modified or C-terminally truncated INSLs and truncated GLP-1Peptide 34 (GPR54), Fertilization promoting peptide (TCP11), Head Activator Neuropeptide (GPR37), Obestatin( GPR39), Neuromedin S, Neuropeptide
S, QRFP-43, PBAN, P518, NPB & NPW, INSL 3 & 7, Endokinin A/B, AF9, BK), receptors (FM-4/TGR-1, NPSR, GPR100, GPR135, GPR142, GPR103, GPR7 & 8, LGR7 & 8, CG6986, NPR-1, PBAN-R) and references.
It has been suggested that several different active conformations of the receptor may exist dependent on the properties of the agonist. Therefore, properly selection with modified peptide ligands for a biased signalling properties may allow the development of drugs that selectively modulate only the therapeutically relevant physiological functions, thereby decreasing the risk of side effects.
 |
Packaged in 96 well plates |
 |
1.5 nanomoles of peptide in 15 micrograms BSA |
|

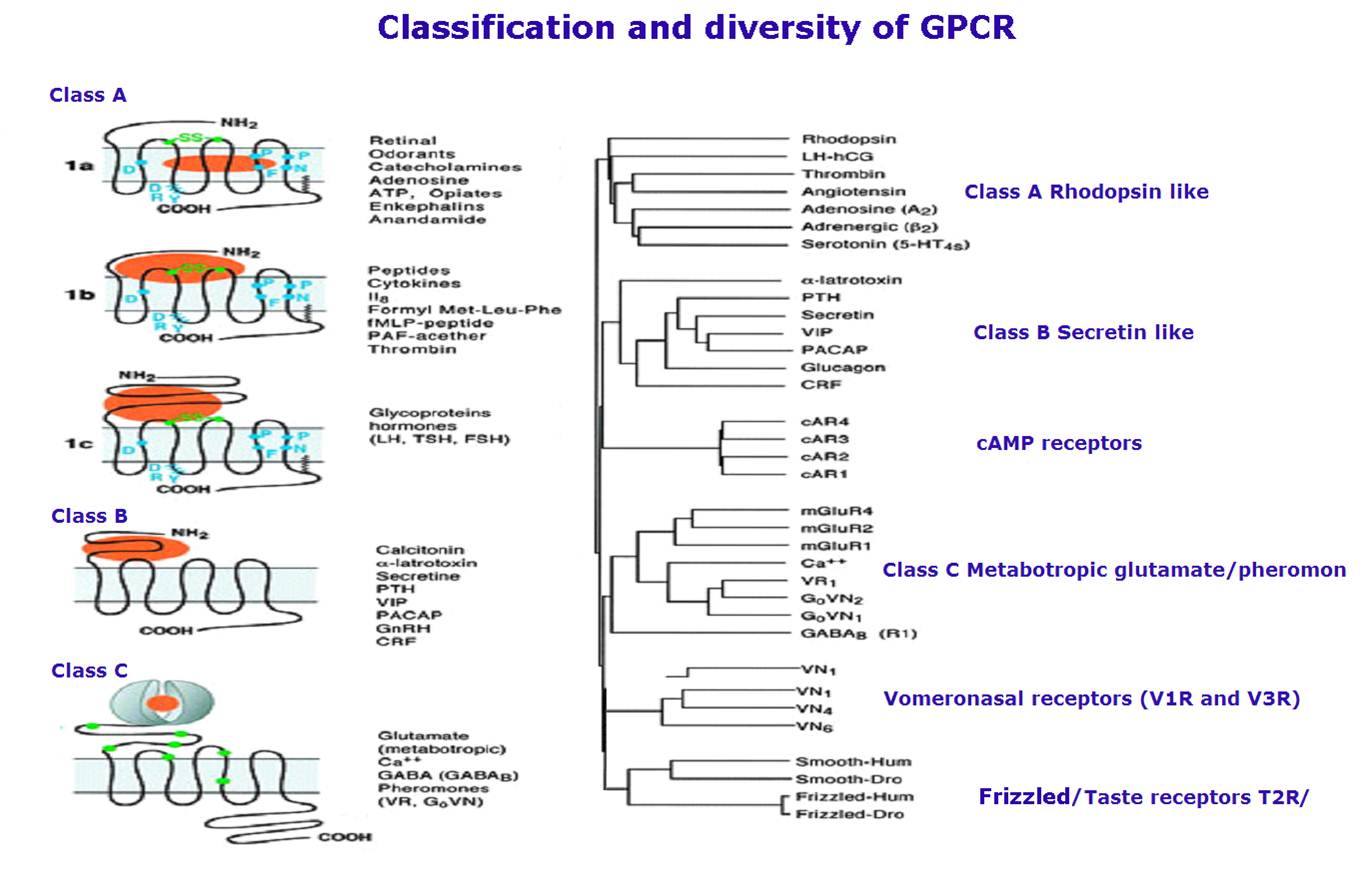
Figure and part of texts from “The EMBO Journal (1999) 18, 1723–1729”.
Three sub-families in “Class A GPCRs” contains being recognized when comparing their amino-acid sequences. Receptors from different families share no sequence similarity. Group 1a contains GPCRs for small ligands and the receptors of the retinal/odorants/opiates/enkephalins. Group 1b contains receptors for peptides whose binding site includees the N-terminal, the the extracellular loops and superior parts of transmembranes. Group 1c contains GPCRs for glycoprotein hormones. “Class B GPCRs” have a similar morphology to group 1c but they do not share any sequence homology. Additional Class includes the family of pheromone receptors(VNs0 and family of “frizzled” and “smoothened (Smo) receptors which involved in embryonic development. Finally, the cAMP receptors (cAR) have only seen found in D. discoideum but its possible expression in vertebrate has not yet been reported.
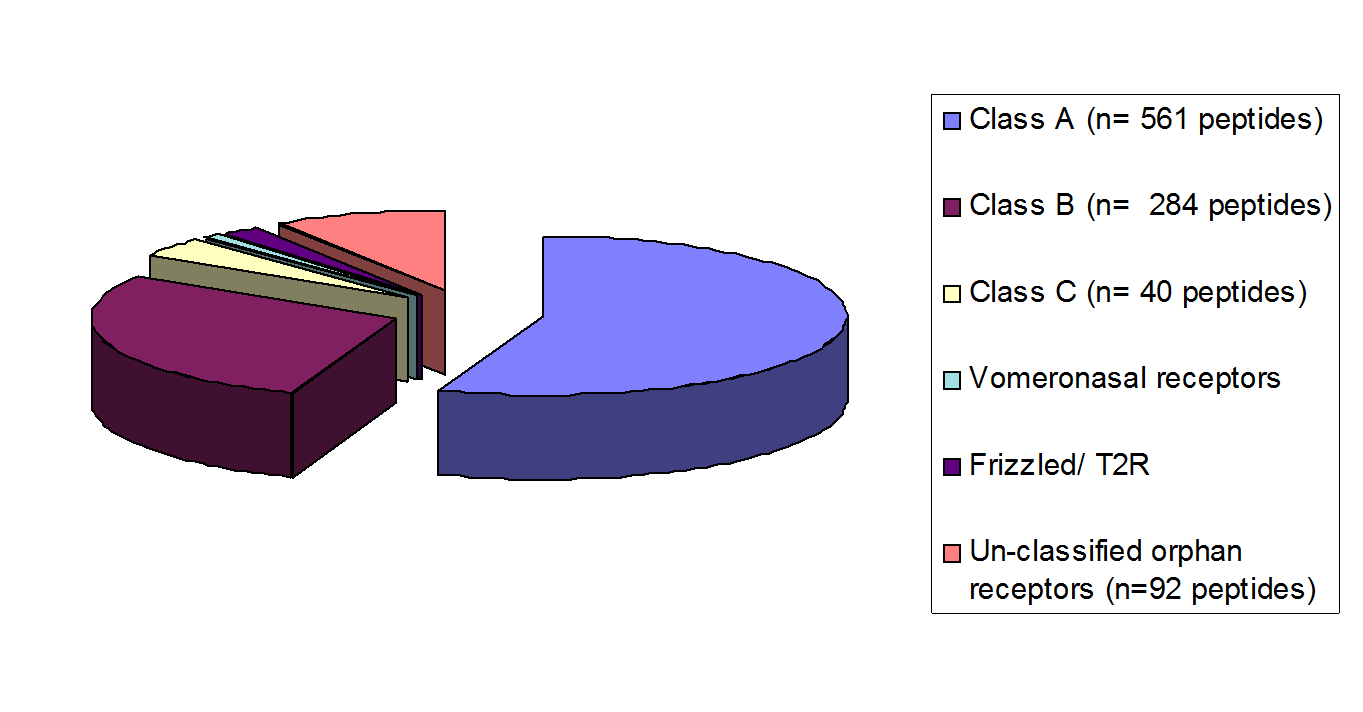
This gPeptide Library (L-004) contains more than 1000 peptides. Each of the peptide has a specific property to interact at least with a known subclass of GPCRs (see the figure on the right). |
 |
|
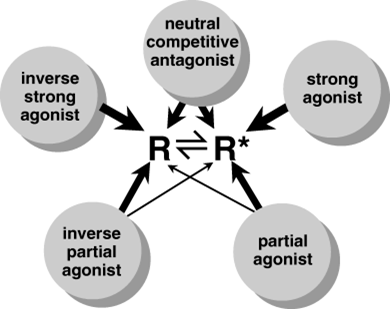
A schematic representation of how the two-state receptor model relates to the action of drugs as strong agonists, partial agonists, neutral competitive antagonists, inverse agonists, and inverse partial agonists. The inactive and active receptor conformations (R and R*, respectively) are in constant equilibrium. |
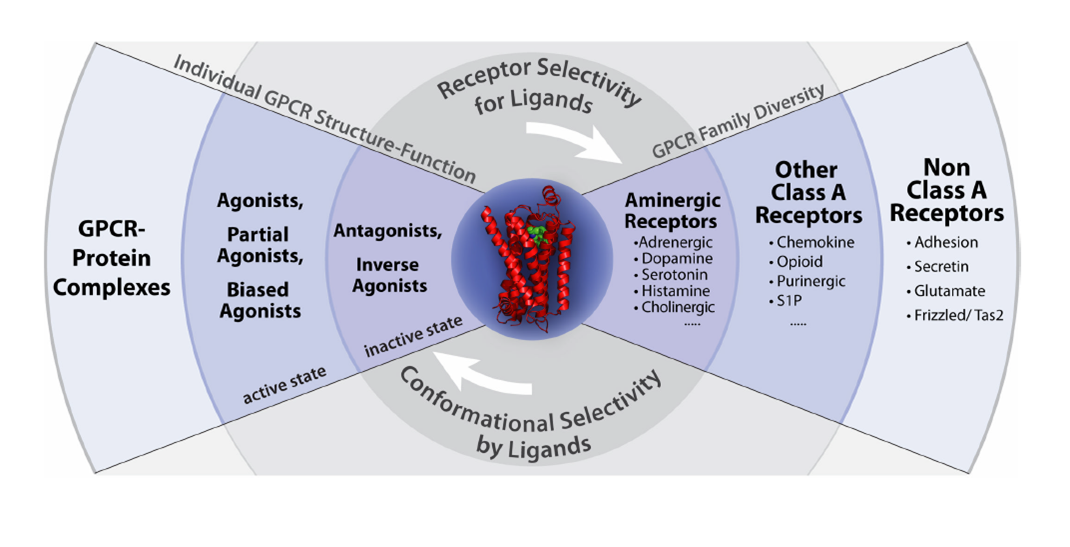
The strategy of the GPCR network includes leveraging each experimentally determined receptor structure towards the understanding of GPCR family diversity and receptor selectivity for ligands (right side) and individual GPCR structure-function and conformational selectivity by ligands (left side). The peptide library is being continuously used to select the best desired function.
Figure from : Raymond C. Stevens et al., GPCR Network: a large-scale collaboration on GPCR structure and function. Nat Rev Drug Discov. 2013 January; 12(1): 25–34.
Want To Discover New Orphan Receptors? Try Phoenix's G Protein-Coupled Receptor Peptide Ligand Library!
More than 50 families of identified or suspected G Protein Coupled Receptor Peptide
Ligands plus its related agonists and antagonists enable a
"reverse pharmacology approach" for large scale screening.
Now over 1000 Peptides, Orphan G Protein Coupled Receptors are included which
are HPLC-purified and fluoresceint free. This library covers various newly identified G Protein Coupled Receptor Ligands, including new Obestatin
(GPR39), RF-amide related peptide QRFP-43, QRFP-26/P518 (GPR103/SP9155/AQ27),
Gln-Alltostatin C(CG7285 & CG13702), Proctolin (CG6986), Endokinin A/B
(NK1), Adipokinetic hormone (ADHR), CCAP (CG6111), PBAN (PBAN-R), H3 Relaxin
(LGR7 & 8), NPB and NPW (GPR7 & GPR8), Icilin (CMR-1), Neuropeptide F (DmNPFR1), RFamide Related Peptides (OT7T022), Neuromedin U
(NmU R-1/FM-3 and NmU R-2/FM-4), human Urotensin II (GPR14), Neuropeptide
AF/FF (NPFF-1 and NPFF-2), Ghrelin (GHS-Rs), Motilin (GPR-38-A), MCH(SLC-1, MCH-R2), Orexins and Hypocretins (HFGAN-72), Prolactin Releasing Peptide (HGR-3/GPR-10/UHR-1), Orphanin FQ (ORL-1) and Apelin(APJ), NEP 1-40 (Nogo-66 receptor)
 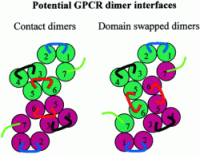
Potential GPCR dimer interfaces. Contact dimers, in which the interface between GPCR monomers involves surface contact between helices of two independent monomers (indicated by color), and domain-swapped dimers, in which helices
6 and 7 are exchanged or "swapped" between GPCR monomers (indicated by color), are illustrated, viewed at the cytoplasmic face of the membrane. Note the origins and locations of the intracellular loops (i1
loop, blue; i2 loop, black; i3 loop, red) contributing to the putative G protein binding surface in the two models.
Breitwieser G.E. Circulation Research. 2004;94:17
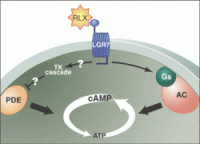
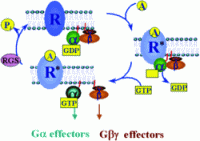
Receptor-mediated G protein activation. The interaction of an endogenous ligand with its cell surface receptor (R) facilitates the coupling of the activate receptor (R*)
with intracellular heterotrimeric G proteins. The R*-G protein coupling promotes the exchange of GDP for GTP on the G-subunit. G-GTP then dissociates from Gß and R*. Both subunits
are free to modulate the activity of a wide variety of intracellular effectors. Termination of the signal occurs when the -phosphate of GTP is removed by the intrinsic GTPase activity of the G-subunit, leaving GDP in the nucleotide binding pocket on G. G-GDP then reassociates with Gß and the cycle is complete. RGS
proteins accelerate the intrinsic GTPase activity of G-subunits, thereby reducing the duration of signaling events.
Cabrera-Vera T. M., et al. Endocrine Reviews 2003, 24 (6): 765-781
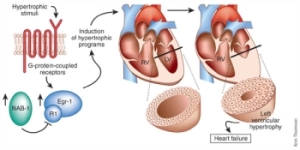
NAB1 negatively regulates pathologic cardiac hypertrophy. Hypertrophic stimuli (such as isoproterenol-phenylephrine or myocardial pressure overload) induce Egr-1 expression, which in turn can
activate pathologic cardiac hypertrophy. These hypertrophic stimuli also increase NAB1, which interacts with Egr-1 through its R1 domain, thereby reducing left ventricular hypertrophy and potentially
cardiac failure. RV, right ventricle; LV, left ventricle.
Levon M Khachigian. Nature Medicine 11, 828 - 829 (2005)

Sarah M. Assmann. G Proteins Go Green: A Plant G Protein Signaling FAQ Sheet. Science, Vol 310, Issue 5745, 71-73 , 7 October 2005

Limited G Protein-Coupled Receptor Peptide list: Click to send E-mail request. |
|
|
L-004
|
|
|


