MMG2-derived peptides (MMG2-DPs)
Myomodulin Gene 2 -derived Peptides
The Aplysia feeding system is advantageous for
investigating the role of neuropeptides in behavioral
plasticity. One family of Aplysia neuropeptides is the
myomodulins (MMs), originally purified from one of the feeding
muscles, the accessory radula closer (ARC). However, two MMs,
MMc and MMe, are not encoded on the only known MM gene. Here,
we identify MM gene 2 (MMG2), which encodes MMc and MMe and
four new neuropeptides. We use matrix-assisted laser
desorption/ionization time-of-flight mass spectrometry to
verify that these novel MMG2-derived peptides (MMG2-DPs), as
well as MMc and MMe, are synthesized from the precursor. Using
antibodies against the MMG2-DPs, we demonstrate that neuronal
processes that stain for MMG2-DPs are found in the buccal
ganglion, which contains the feeding network, and in the
buccal musculature including the ARC muscle. Surprisingly,
however, no immunostaining is observed in buccal neurons
including the ARC motoneurons. In situ hybridization reveals
only few MMG2-expressing neurons that are mostly located in
the pedal ganglion. Using immunohistochemical and
electrophysiological techniques, we demonstrate that some of
these pedal neurons project to the buccal ganglion and are the
likely source of the MMG2-DP innervation of the feeding
network and musculature. We show that the MMG2-DPs are
bioactive both centrally and peripherally: they bias egestive
feeding programs toward ingestive ones, and they modulate ARC
muscle contractions. The multiple actions of the MMG2-DPs
suggest that these peptides play a broad role in behavioral
plasticity and that the pedal-buccal projection neurons that
express them are a novel source of extrinsic modulation of the
feeding system of Aplysia.
Proekt A, et al. J Neurosci. 2005 Oct 19;25(42):9637-48.

MMG2 precursor protein. A signal peptide is
shown by the white rectangle. Basic amino acids (K and R) are
shown in bold. These are potential processing enzyme cleavage
sites. Amidated peptides detected by MALDI (Fig. 4) are shown
by the gray rectangles: MMc, MMe, and four novel peptides
(MMG2-DPa, MMG2-DPb, MMG2-DPd, and MMG2-DPf). Peaks
corresponding to the underlined nonamidated connecting
peptides (pMMG284 –112, 3091 Da; pMMG2131–168, 4061 Da;
pMMG2171–199, 3231 Da) were also detected by MALDI (Fig.
4)
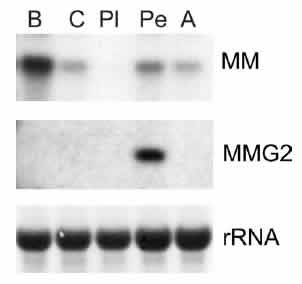
Comparison of expression of MM and MMG2 mRNAs. Northern
blots with either MMor MMG2 probes using RNA extracted from
buccal (B), cerebral (C), pleural (Pl), pedal (Pe), and
abdominal (A) ganglia are shown. Ribosomal RNA (rRNA) bands
are shown to demonstrate that the same amount of RNA was
loaded into each lane.
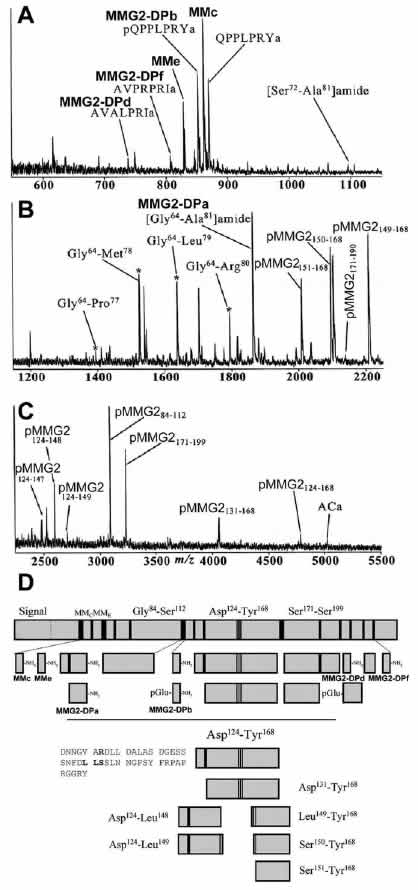
MALDI analysis of individual pedal neurons reveals MMG2
precursor processing. A representative mass spectrum of a
single Aplysia pedal neuron is shown. The mass scale is
divided into low (m/z 550 –1150) (A), middle (m/z 1200 –2250)
(B), and high (m/z 2450–5500) (C) mass ranges. Mass spectral
peaks were assigned based on the observed mass and the
sequence of MMG2. D, Summary of the MMG2 precursor processing.
Solid vertical lines represent cleavage sites, with black bars
indicating dibasic sites that are entirely cleaved. Lightly
shaded blocks represent peptides detected by MALDI. The
processing of connecting peptide Asp 124-Tyr 168, highlighted
below, shows numerous novel processing sites involving Leu-Leu
and Leu-Ser cleavages.


